Polymerase Chain Reaction, Steps of PCR, Restriction enzymes and Cystic Fibrosis
Diagnostic techniques in molecular biology are techniques concerned with genes: the DNA or RNA or their products (proteins). The first step is the extraction of the nucleic acids from the cell: DNA, and RNA.
Content
- PCR: for amplification of a target sequence.
- Gel electrophoresis: to visualize bands and analyze them.
- Restriction enzymes (RFLP).
- Hybridization and Blotting techniques:
Amplification of a target sequence:
- Cloning (recombinant DNA, chimeric DNA, hybrid DNA): The target gene will replicate in a host cell using its enzymes and all control systems.
- PCR: cell-free amplification, there is no host cell, the replication of the DNA occurs in a test tube, in an apparatus.
- The final result will be a huge amount of DNA.
- In cloning, as a host cell is used and it has protein synthesis machinery, we can produce a protein that can be used for therapy (insulin) or vaccine (HB).
Application
- To detect infectious agents (HIV, HCV, HCB).
- Diagnosis of different genetic diseases: thalassemia, sickle cell anemia.
- For genetic prenatal diagnosis. (cells from the amniotic fluid or chorionic villi).
- To establish precise tissue types for transplantation.
- In research: understand the molecular basis of diseases (cancer) and study evolution.
- Forensic analysis of DNA samples. DNA isolated from a single hair is sufficient to determine whether the sample comes from a specific individual.
- Gene therapy: genetic engineering.
- By cloning: production of proteins that can be used as drugs (insulin. GH) or as vaccines (HB).
According to the type of mutation or disease needed to be diagnosed or studied:
- PCR → gel electrophoresis
- PCR → Add restriction enzymes → gel electrophoresis
- The sample + RE → gel electrophoresis → blot technique (hybridization with a probe)
Polymerase Chain Reaction (PCR)
Polymerase Chain Reaction is a rapid and sensitive method of amplifying a target sequence of DNA, The method can be used to amplify DNA sequences from any source: bacterial, viral, plant or animal.
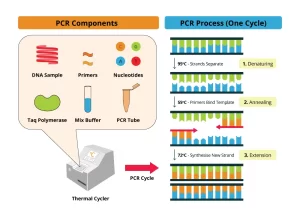
Required Components of PCR
Steps of PCR
- The DNA sample is first heated to separate the 2 strands (Denaturation).
- Two oligonucleotide primers are added and allowed to hybridize to complementary sequences on opposite strands of DNA and flank the target sequence.
- Each strand of DNA is copied by a DNA polymerase starting at the primer site, The two DNA strands each serve as a template for the synthesis of new DNA from the 2 primers.
- Repeated cycles result in the amplification of DNA segments of defined length. (ex. 20 cycles provide an amplification of 106).
Overview of PCR
- Temperature Cycling: Denaturation ( 94°), Annealing (55°), and Extension (72°).
- Every cycle of DNA between primers is duplicated.
Reverse transcriptase PCR
- In reverse transcriptase PCR, mRNA is detected.
- Ex: HIV RNA in blood can be detected as early as 4 weeks after infection.
Real-time PCR
Quantitation of the number of viruses present in a sample can be calculated, ex: viral load in HIV or HBV, So the treatment modalities can be planned and the response to treatment can be assessed.
Gel Electrophoresis
In electric current, The DNA fragment will migrate according to its size towards the positive electrode because DNA contains negatively charged phosphate groups, So, DNA segments can be separated, and Agarose or polyacrylamide gel can be used.
Restriction fragment length polymorphism (RFLP)
- Polymorphism is detected by using restriction enzymes.
- Polymorphism: more than one form:
Restriction enzymes
- Endonuclease that cuts specific sequences of the DNA.
- Bacterial in origin: protection from viral infection.
- Used in molecular biology techniques to cut DNA into fragments.
- In the presence of a point mutation, the site of action of the enzyme (restriction site) can be abolished or created.
- Thus using the restriction enzyme in the DNA target gene, gives different results in case of mutation. for each gene, there are 2 copies.
Principle
- Isolation of DNA.
- DNA is amplified by PCR.
- Amplified DNA is incubated with restriction endonucleases.
- Then electrophoresis.
- Visualization of different bands.
Hybridization and blot techniques
- Hybridization means specific reassociation between complementary strands of nucleic acids (DNA&DNA, DNA&RNA, or RNA&RNA).
- A probe is a single-stranded piece of DNA, Labeled (either with radio-isotope or with non-radioactive label). Its nucleotide sequence is complementary to the target DNA.
Blotting techniques
Blotting analysis can separate and detect specific nucleic acid sequences or proteins in complex mixtures.
Types
- Southern blotting: detects DNA with labeled DNA probes.
- Northern blotting: detects RNA with labeled DNA probes, It is useful in determining whether a specific mRNA is expressed in a particular tissue.
- Western blotting: detects proteins by using labeled antibodies.
Principle
- Extraction of DNA.
- DNA is cleaved with a restriction enzyme.
- Agarose gel electrophoresis.
- Denature the DNA into single strands (by alkali).
- Transfer to nitrocellulose membrane (blotted).
- Hybridize with a labeled probe.
- Band visualization. The labeled probes detect specific DNA sequences.
Amplification of DNA is done by either: Recombinant DNA, or PCR.
Analysis of DNA can be made through: Electrophoresis only, R.E. & Electrophoresis (RFLP), R.E. & Electrophoresis & blot & probe. (Southern blot).
Clinical applications
Sickle Cell Disease
- Sickle cell anemia is a disease of red blood cells, It is caused by a mutation in the hemoglobin gene, A single base change results in a single amino acid substitution.
- Sickle cell anemia is considered a recessive trait since both chromosomes have to cany the mutation in order for the full-blown disease symptoms to appear.
- The sickle cell mutation eliminates a restriction enzyme site – the recognition site for the restriction enzyme MstII.
- To detect the sickle cell mutation, a patient’s DNA is digested with MstIl, and a Southen blot is performed using a probe corresponding to this region of the hemoglobin gene.
- The presence or absence of the sickle cell mutation can be determined based on the size of the fragment identified by the probe.
Cystic Fibrosis
- The cause of the disease appears to be a mutation in the gene encoding, the cystic fibrosis transmembrane conductance regulator (CFTR), a membrane protein involved in transporting ions across epithelial surfaces, such as the linings of the lungs and intestines.
- Cystic fibrosis is caused by the deletion of 3 DNA bases.
- Missing codon in mRNA.
- Amino acid phenylalanine therefore missing from transmembrane protein in the lungs.
- This mutation is detected by sequence analysis of PCR-amplified DNA or by hybridization with mutation-specific probes.
You can subscribe to Science Online on YouTube from this link: Science Online
You can download the Science Online application on Google Play from this link: Science Online Apps on Google Play
Cloning of DNA sequences, Importance of recombinant DNA & Uses of human genome
Molecular technology (Genetic Engineering) advantages, Techniques and uses
Importance of Nucleosides, Nucleotides, Purines, Pyrimidines & Sugars of nucleic acids
Regulation of the cell cycle, DNA synthesis phase, Interphase & Mitosis