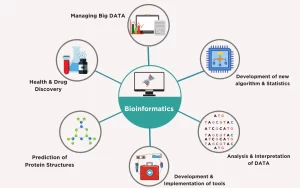
Bioinformatics applications in diagnosis of disease, uses of the sequenced DNA
Bioinformatics is a scientific subdiscipline that involves using computer technology to collect, store, analyze, and disseminate biological data and information, such as DNA and amino acid sequences or annotations about those sequences, Bioinformatics involves biologists who learn programming, or computer programmers, mathematicians or database managers who learn the foundations of biology.
What is bioinformatics?
Bioinformatics is the collection, classification, storage, and analysis of biochemical and biological info using computers especially as applied to molecular genetics and genomics, Bioinformatics allows us to handle the huge amounts of data involved, It involves processing, storing, and analyzing biological data, Which might include creating databases to store experimental data, predicting the way that proteins fold up, Modelling how all the chemical reactions in a cell interact with each other.
Bioinformatics combines the following fields to analyze biological data:
- Computer science.
- Statistics.
- Molecular biology.
Applications of bioinformatics in molecular biology
- Identification of gene.
- Prediction of gene expression and protein–protein interactions.
- Diagnosis of genetic disorders of different diseases.
- Analysis of mutations.
- Drug design and discovery.
- Prediction of protein structure.
All this information is supplied through databases. Due to the huge amount of available genetic information, there is a huge number of available databases covering almost everything regarding nucleic acids‘ and proteins‘ sequences, molecular structures, and phenotypes. These databases are called biological databases.
Therefore, biological databases include: an organized collection of biological data, collected from scientific experiments, published literature, and computational analysis, Research areas include genes, proteins, metabolic pathways, gene expression, and disease.
Types of Biological Databases
- Sequence and structure databases: eg. Genbank database.
- Molecular functions: eg. Transcriptional regulation (TRANSFAC databases).
- Biological processes: eg. Metabolic pathways (KEGG database).
BLAST
The National Center for Biotechnology Information (NCBI) is one of the largest biological databases present online (http://www.ncbi.nlm.nih.gov). In NCBI, BLAST (Basic Local Alignment Search Tool) can be found, BLAST is a very useful tool for comparing primary biological sequence information, such as the amino-acid sequences of proteins or the nucleotides of DNA sequences.
BLAST finds regions of similarities and differences between nucleotides or amino acids entered by the user and those stored in the database (wild type or normal), and calculates the percentage of similarity, It also helps in the prediction of which gene does a certain nucleotide sequence belongs to, and also which protein does a certain amino acid sequence belongs to.
There are now a handful of different BLAST programs available, These different programs vary in query sequence input, the database being searched, and what is being compared, These programs include:
- Nucleotide-nucleotide BLAST (blastn): The user enters a DNA query (nucleotide sequence), and blastn shows the most similar gene sequence from the stored genome database.
- Protein-protein BLAST (blastp): The user enters a protein (amino acid sequence) query, and blastp shows the most similar protein from the stored protein database.
- Nucleotide translation-protein (blastx): This program compares the translated nucleotide sequence (both strands) against a protein sequence database.
- Protein-nucleotide 6-frame translation (tblastn): This program compares a protein to nucleotides (user enters amino acid sequence and (blast shows the proposed corresponding nucleotides), Of these programs, BLASTn and BLASTp are the most commonly used because they use direct comparisons, and do not require translations.
Example of application of bioinformatics in diagnosis of disease
Clinical case:
- A mother brought her child to the emergency room because the child was complaining of difficulty in breathing.
- On examination, the child was a 13-year-old male, on a wheelchair and his limbs seemed to be much thinner than they should be for his age.
- The calf muscles however appeared to be larger than normal.
- On history taking, the mother reported that her child was born apparently normal. However, she noticed that he had delayed motor milestones (like sitting, standing, and walking).
- During his childhood, the boy always had difficulty in climbing stairs, moving, and even difficulty in getting up from the floor when he fell down!
- She added that at the age of 12, moving was almost impossible for her son so she had to use the wheelchair.
- Investigations made by the doctor revealed highly elevated serum creatine kinase levels indicating muscle tissue damage and muscle biopsy revealed atrophic muscle tissue.
- The doctor suspected the child was suffering from Duchenne Muscle dystrophy and resorted to DNA sequencing to confirm his diagnosis.
What are the uses of the sequenced DNA?
- Comparing genome: Once the patient’s nucleotide sequence is obtained, it can be entered in BLASTn, and BLASTn will show genes of the closest similarity with this sequence, and will point out the differences.
- Translation.
- Translate sequence into protein.
- Protein shape: Determine the 3D structure of the protein.
Steps of diagnosis using bioinformatics
- Identify the suspected mutated gene: in this case, the suspected mutation is in the “dystrophin” gene, which is located on the short arm of the X chromosome. Dystrophin is responsible for connecting the cytoskeleton of each muscle fiber to the underlying basal lamina (extracellular matrix), through a protein complex containing many subunits.
- Isolate the desired sequence of the suspected gene by PCR, then identify the nucleotide sequence of the PCR product using the sequencer.
- Enter the nucleotide sequence obtained from sequencing into the nucleotide BLASTn designed area, choose the organism you are studying, and then press blast.
- BLAST will align your sequence with the sequences in different genes stored in the database and give out the options. Choose the one with the highest percentage of similarity with your sequence. We can analyze this result by saying that the sequence of the patient was identified on the X chromosome, and the similarity between it and the stored normal sequence on the database is 99%. This is due to the deletion of 3 bases on the patient’s gene leading to mutation.
- In order to translate this sequence to protein we can use EXPASY (Expert Protein Analysis System, http://web.expasy.org/translate/). The results will appear as follows. Choose the frame with the longest genetic message starting with methionine and ending with stop.
- Copy this amino acid sequence and return to Blastp, to identify which protein is the most similar to your obtained amino acid sequence.
- Compare the similarity of your protein with the protein database in BLASTp. As shown, the patient protein has the highest similarity with dystrophin, however, the deletion mutation in the gene resulted in a protein that is only 87% identical to the original protein.
The 3-dimensional structure of the protein can be obtained by further entering the amino acid sequence in Cn3D (http://www.ncbi.nlm.nih.gov/Structure/CN3D/en3d.shtml).
To summarize the results:
- The patient’s nucleotide sequence is located on chromosome X.
- Identity: 99% with a normal gene.
- Mutation due to bases deletion.
- Mutation in DNA resulted in mutated protein with only 87% similarity to the normal protein (dystrophin).
- Abnormal structure of dystrophin resulted in clinical features of Duchenne muscle dystrophy.
You can subscribe to Science Online on YouTube from this link: Science Online
You can download the Science Online application on Google Play from this link: Science Online Apps on Google Play
Cloning of DNA sequences, Importance of recombinant DNA & Uses of human genome
Molecular technology (Genetic Engineering) advantages, Techniques and uses
Importance of Nucleosides, Nucleotides, Purines, Pyrimidines & Sugars of nucleic acids
Regulation of the cell cycle, DNA synthesis phase, Interphase & Mitosis